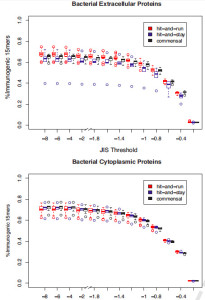
JIS by bacterial group. JIS distributions of peptides separated by both extracellular (top) vs. cytoplasmic (bottom) and by bacterial group (colors within each plot). Box plots are as described for Fig. 2. The striking outlier in top panel with much lower %immunogenic 15mers than others is a hit-and-stay bacterium Mycobacterium tuberculosis Oshkosh.
Graduate student Lu He, working with EpiVax CEO De Groot and collaborator Chris Bailey Kellog, recently reported that bacteria ‘copy and paste’ sequences from the human genome to escape immune response. She calls this escape mechanism “bacterial immune camouflage“.
She used the JanusMatrix tool to develop a new measure of immune escape, the Janus Immunogenicity Score (JIS), to assess the likelihood of immune recognition of a bacterial antigen. As shown at right, Lu He found that certain types of bacteria that persist inside of humans (“hit-and-stay”, such as Herpes, EBV, and CMV) seem to use this mechanism in order to escape immune response, much more so than those bacteria that cause short-lived infection (“hit-and-run”, such as Ebola and Marburg). Tuberculosis was strikingly different from all other bacteria, an observation that deserves further investigation.
The JanusMatrix score has previously been shown to be predictive of “suppressive” vs. “activating (T regulatory vs. T effector, respectively) in experimental data, and in a previously published study, was also able to distinguish viruses representative of the hit-and-stay vs. hit-and-run phenotypes.
In the new study, JanusMatrix was used to evaluate genomic differences among representative hit-and-run, hit-and-stay, and commensal bacteria. Overall, extracellular proteins were found to have different JIS profiles from cytoplasmic ones. The analysis points to a new way to explain bacterial immune camouflage or escape, and suggests that the JanusMatrix approach could be used to select and optimize of candidates for vaccine design.
