An Update on EpiVax’ H7N9 “FastVax” Progress
EpiVax scientists are currently in discussions with groups in China, Canada and the US about producing a vaccine. Here we highlight a few key points about the current progress of EpiVax’ FastVax vaccine design:
(1) While H7N9 influenza may be mutable, selecting highly conserved and immunogenic components for vaccines is our expertise. Some aspects of the genome may not be conserved, and are ‘extra information’. Basically, all living organisms have to have some key regions of their genome that enable them to survive. A simple analogy: people need their thumbs. They can change their hair color, but they can’t live without their thumbs (it makes us human). Or, you could say, we’re going to make a vaccine that includes thumb-like, immutable but very immunogenic components. These components are called epitopes.
(2) That said, the sequence of H7N9 indicates that it will be hard to design a vaccine for this virus using traditional methods (based on HA). Our in-house experts, using our immunogenicity screening tools, have predicted that HA will be a poor immunogen. Compared to H1 and H3 (two of the current circulating strains of influenza), H3 hemagglutinin (HA), has far fewer T cell epitopes. See figure below for a comparison of the three influenza HA’s.
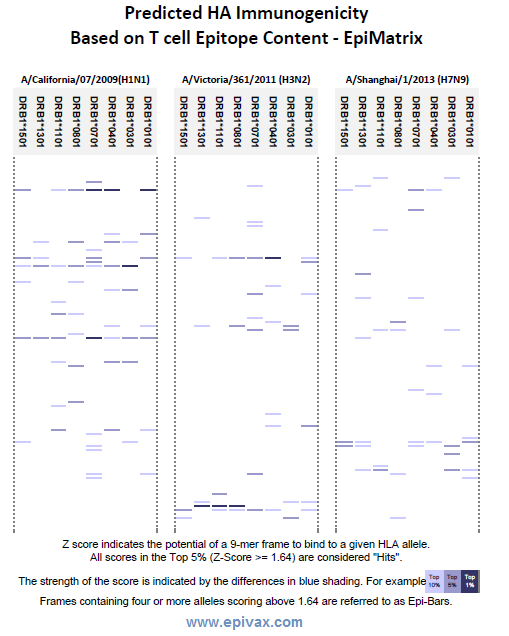
This graphic shows an EpiMatrix analysis of flu HA (H1, H3, H7). The blue bars represent T cell epitopes; the darker the bar, the stronger the epitope. A larger quantity of bars correlates with higher immunogenicity of the proteins. More Info.
(3) We suspect that the lack of T cell epitopes in the HA protein of H7N9 will reduce effective antibody response. It takes T-help to make high titer, high affinity IgG antibodies. Lack of good T cell epitopes, and (as shown above) very poor conservation of T helper epitopes in H7 as compared to H3 and H1, suggests that making a vaccine based on HA alone might not be the best approach.
In the figure above, we have provided a very preliminary snapshot of our influenza HA protein immunogenicity analysis, comparing recent strains of circulating H1 virus HA protein with that of H3, and emergent H7 for T cell epitope content. Note that – not only do the epitopes not align, but there are far fewer in H7, suggesting very low immunogenicity of HA.
For more information on EpiVax Immunogenicity Screening for Vaccines and Protein Therapeutics – see the following links: Vaccine design and protein therapeutics. For our publications on vaccine research, see here. For information about our academic collaboration with the Institute for Immunology and Informatics, click here. For a list of our scientific collaborators, click here.
Sign up for EpiVax’s Newsletter to be kept up-to-date on our progress – Sign Up Here!
For blogs that we follow and links to flu-relevant publications, see below:
- Link to: https://www.promedmail.org/
- Link to https://crofsblogs.typepad.com/
- https://www.uq.edu.au/vdu/VDUInfluenza_H7N9.htm
- https://afludiary.blogspot.com/
- https://www.cidrap.umn.edu/index.html
- https://www.who.int/csr/don/2013_04_15/en/index.html
Referenced journal articles:
- Uyeki TM, Cox NJ. Global Concerns Regarding Novel Influenza A (H7N9) Virus Infections. N Engl J Med. 2013 Apr 11. [Epub ahead of print] PubMed PMID: 23577629.
- Skowronski DM, Janjua NZ, De Serres G, Winter AL, Dickinson JA, Gardy JL, Gubbay J, Fonseca K, Charest H, Crowcroft NS, Fradet MD, Bastien N, Li Y, Krajden M, Sabaiduc S, Petric M. A sentinel platform to evaluate influenza vaccine effectiveness and new variant circulation, Canada 2010-2011 season. Clin Infect Dis. 2012 Aug;55(3):332-42. doi: 10.1093/cid/cis431. Epub 2012 Apr 26. PubMed PMID: 22539661.
- Schanen BC, De Groot AS, Moise L, Ardito M, McClaine E, Martin W, Wittman V, Warren WL, Drake DR 3rd. Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4(+) T cells against the swine-origin H1N1 influenza virus. Vaccine. 2011 Apr 12;29(17):3299-309. doi: 10.1016/j.vaccine.2011.02.019. Epub 2011 Feb 22. PubMed PMID: 21349362; PubMed Central PMCID: PMC3130614.
- Ellebedy AH, Ducatez MF, Duan S, Stigger-Rosser E, Rubrum AM, Govorkova EA, Webster RG, Webby RJ. Impact of prior seasonal influenza vaccination and infection on pandemic A (H1N1) influenza virus replication in ferrets. Vaccine. 2011 Apr 12;29(17):3335-9. doi: 10.1016/j.vaccine.2010.08.067. Epub 2010 Sep 15. PubMed PMID: 20840835; PubMed Central PMCID: PMC3026885.
- De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008-2009 conventional influenza vaccine. Vaccine. 2009 Sep 25;27(42):5740-7. doi: 10.1016/j.vaccine.2009.07.040. Epub 2009 Aug 4. PubMed PMID: 19660593.
- De Groot AS, Martin W, Moise L, Guirakhoo F, Monath T. Analysis of ChimeriVax Japanese Encephalitis Virus envelope for T-cell epitopes and comparison to circulating strain sequences. Vaccine. 2007 Nov 19;25(47):8077-84. Epub 2007 Sep 29. PubMed PMID: 17942198.
- Weber CA, Mehta PJ, Ardito M, Moise L, Martin B, De Groot AS. T cell epitope: friend or foe? Immunogenicity of biologics in context. Adv Drug Deliv Rev. 2009 Sep 30;61(11):965-76. doi: 10.1016/j.addr.2009.07.001. Epub 2009 Jul 18. Review. PubMed PMID: 19619593.
- De Groot AS, McClaine E, Moise L, Martin W. Time for T?: Thoughts about the 2009 novel H1N1 influenza outbreak and the role of T cell epitopes in the next generation of influenza vaccines. Hum Vaccin. 2010 Feb;6(2):161-63. Epub 2010 Feb 19. PubMed PMID: 20431339; PubMed Central PMCID: PMC2936654.
Referenced Websites:
- WHO, Human infection with influenza A(H7N9) virus in China – update, 12 April 2013. https://www.who.int/csr/don/2013_04_12/en/index.html
- H7N9 genetic analysis raises concern over pandemic potential, Lisa Schnirring Staff Writer, Apr 12, 2013 (CIDRAP News), https://www.cidrap.umn.edu/cidrap/content/influenza/avianflu/news/apr1213genetic.html
- Avian Flu Diary Blog, 12 April, 2013. https://afludiary.blogspot.com/search?q=Tashiro&max-results=20&by-date=true
- Global Initiative on Sharing All Influenza Data (GISAID), https://platform.gisaid.org/epi3/frontend#22a3ff
- iCubed HA Analysis with Epimatrix.https://www.immunome.org/news/ivax/epimatrix-analysis-of-a-complete-set-of-h1n1-hemagglutinin-peptides/
- EpiVax Blog, April 12, 2013, https://www.epivax.com/blog/h7n9-shanghai-2013-the-new-stealth-virus/
- EpiVax Blog, April 7, 2013, https://www.epivax.com/blog/shanghai-2013-extremely-rapid-h7n9-vaccine-design-by-epivax/

