EpiVax is celebrating 25 years of innovative immunoinformatics tool development. The company’s mission is to improve the safety and efficacy of vaccines and protein therapeutics, while working to improve human health everywhere.
1998  EpiVax Founded by Dr. Annie De Groot and Mr. Bill Martin
EpiVax Founded by Dr. Annie De Groot and Mr. Bill Martin



Following years of training in internal medicine, immunology, immunoinformatics, and vaccinology, Annie De Groot MD joined the faculty of the Brown University Medical School in 1993 and founded the TB/HIV Research Laboratory where the EpiMatrix technology was developed. This was right at the time when the first full bacterial genome was sequenced (by a former Brown University colleague, Rob Fleischman). Recognizing the need to develop in silico tools to address the volume of pathogens that were going to be sequenced in the coming years, and Annie decided to start a company. But she needed programming expertise to make it happen. In typical Rhode Island fashion Annie met “a friend of a friend” with expertise, Bill Martin. Annie and Bill worked together on developing a ‘web-based epitope-mapping tool’ for the NIH, and then proposed to spin out a company, with support from Slater Technology Fund a Rhode Island business incubator. EpiMatrix technology was licensed to the company from Annie’s lab at Brown University in 1998. Edgy since inception, the company has been living up to its motto: “Science Without Fear.”
2005  HIV and TB Vaccine Immunogenicity Proof of Concept
HIV and TB Vaccine Immunogenicity Proof of Concept
Having established the company, EpiVax team members set to work. Their initial focus was on HIV, the causative agent of the AIDS epidemic. Work in TB and HIV was funded by NIH and Gates foundation, in collaboration with the Sequella Foundation’s Carol Nacy and Leo Einck. EpiMatrix and complimentary immunoinformatic algorithms were used to screen the genomes of the causative agents of tuberculosis and AIDS and to design T cell epitope-driven vaccine candidates. Solid, meaningful research collaborations were also established in Mali to partner with scientists working in countries most affected by these pathogens. Epitopes were validated using samples from infected humans, and when tested in humanized HLA-DR transgenic mice, the vaccine candidates proved to be effective. These data confirmed the utility of immunoinformatics tools to design innovative vaccines. EpiVax’s HIV and TB vaccine research was primarily grant funded and continues to be a focus of interest when funding and collaborations are available.
2006  ISPRI: Interactive Screening Protein Reengineering Interface
ISPRI: Interactive Screening Protein Reengineering Interface
The Epi-Team applied their knowledge of computational vaccinology to produce a web-based toolkit, ISPRI, to screen for the safety (immunogenicity) of biologics, like protein therapeutics and antibodies. ISPRI was developed with an enhanced version of the EpiMatrix technology at its core, and complimentary algorithms (ClustiMer, BlastiMer, OptiMatrix) were included, making ISPRI a key platform for use in the biologics discovery process. Gene Koren (Genentech/Amgen), Andy Glasebrook (Lilly), and Tony Mire Sluis (FDA) were early supporters of the ISPRI approach. Since its creation, the ISPRI platform has been utilized for the screening of millions of biologic sequences in both internal EpiVax analyses and external analyses performed by licensees of the platform from the largest pharmaceutical companies in the world.

2008  Tregitope Discovery
Tregitope Discovery
While screening antibodies for immunogenicity, the EpiVax team identified T cell epitopes that were highly conserved across IgG molecules. Upon further investigation, the team found and validated that these conserved sequences naturally induced regulatory T cells that modify, or dampen, the immune response. This discovery of “Tregitopes” contributed to the concept that there are regions of molecules that can “turn-off” the immune response. Tregitopes enhanced EpiVax’s ability to accurately predict and modulate immunogenicity. They also paved the way for another important development, the JanusMatrix tool.

2009  iVAX: A Web-Based Vaccine Design Platform
iVAX: A Web-Based Vaccine Design Platform
As interest in licensing the ISPRI platform for screening biologics took off, NIH funding materialized to support the production of a web-based toolkit, iVAX, geared for rapid T cell epitope-driven vaccine design. As with ISPRI, the iVAX toolkit was based upon the EpiMatrix technology, and complimentary algorithms such as Conservatrix, EpiAssembler, VaxCAD were included, making iVAX a seamless vaccine design work environment that could be accessed via the web. “iVAX Toolkit” training sessions attracted international vaccine collaborations such as Dan Hoft and Chris Eickhoff (SLU), Yoshimasa Takahashi (Japan) and Horacio Lara (Mexico), among many others. Since its development, the iVAX vaccine design platform has been utilized in the design of all EpiVax vaccine programs (and those of many collaborative partners), including biodefense/infectious disease and cancer research. Publications can be found here.

2009  TulyVax Efficacy Proof of Concept
TulyVax Efficacy Proof of Concept
Francisella tularensis, the causative agent of Tularemia, is one of the most infectious bacterial pathogens known. Because of its high infectivity and mortality rate, the aerosolized form of the bacteria represents a potentially dangerous biological weapon. As there are still no vaccines currently approved for public use, EpiVax developed a research program to identify a vaccine candidate. In this research, the EpiVax team constructed a T cell epitope-driven subunit vaccine candidate, TulyVax. It was shown that the vaccine candidate increased the rate of survival in humanized HLA-DR transgenic mice challenged with a lethal dose of the pathogen. These results demonstrated the efficacy of a T cell epitope-based vaccine and suggested that such an approach might be widely applicable to the development of vaccines for bacterial pathogens. This was but one of several ‘gene to vaccine’ efforts that resulted in successful proof of concept in pre-clinical challenge models. Others include VennVax and, most recently, a COVID-19 vaccine, EPV-COV-19 (see below). Lenny Moise, PhD, directed these efforts at EpiVax until 2022, and remains active in the field.
2009  Role of T memory in Pandemic H1N1 Influenza Immune Response
Role of T memory in Pandemic H1N1 Influenza Immune Response
During the emergence of the pandemic H1N1 strain of influenza in 2009, the virus was associated with unique age-related susceptibility, with higher incidence of illness and death among young persons and lower incidence among older persons. Since pre-existing H1N1 antibodies were not cross-reactive with the prior seasonal vaccine, EpiVax hypothesized that response to T cell epitopes that were cross-conserved between pandemic H1N1 and the 2008 seasonal influenza strains may have contributed to partial protection from the illness among older adults. Using immunoinformatics tools, EpiVax identified the regions cross-conserved between the two strains and showed that vaccination with the conserved influenza T cell epitopes lowered viral load in humanized HLA-DR transgenic mice challenged with the pandemic H1N1 strain. This study highlighted the importance of T cell epitope cross conservation and induction of T memory by those cross-reactive epitopes, as a lever for building better vaccines.

2011  VennVax Efficacy Proof of Concept
VennVax Efficacy Proof of Concept
The potential for smallpox to be disseminated in a bioterror attack prompted development of new, safer, smallpox vaccination strategies. EpiVax designed and evaluated immunogenicity and efficacy of a T cell epitope-driven vaccine candidate, VennVax, based on conserved vaccinia/variola viral sequences identified using the iVAX toolkit. Vaccination in humanized HLA-DR transgenic mice provided significant protection, as 100% of vaccinated mice survived when challenged with a lethal dose of the virus. This demonstrated that protective immunity to smallpox does not require priming with an antibody response-inducing vaccine.

2013  JanusMatrix: Assessing Immune Camouflage
JanusMatrix: Assessing Immune Camouflage
As the EpiVax team continued to delve into vaccine design research for various pathogens, the scientists questioned how some pathogens, like HIV, HSV, and HCV, can evade the human immune response. Guided by their previous Tregitope discovery, the EpiVax team found that pathogens in fact evolve their proteins to contain sequences that look like human Tregitopes to camouflage themselves from the human immune response by appearing as “self”. To assess the cross-conservation of pathogens with the human proteome, EpiVax scientists developed a new algorithm, JanusMatrix. The utility of JanusMatrix has been demonstrated in multiple studies, which have validated that pathogen sequences with extensive human cross-conservation can evoke a regulatory (or dampening effect on the) immune response. This new tool, now applied to both vaccine design and biologics screening, has advanced EpiVax’s in silico immunogenicity prediction even further ahead of other available bioinformatics tools, and it has been particularly key in EpiVax’s most recent vaccine design research programs.

2016  EpiVax Therapeutics: The Ancer to Cancer
EpiVax Therapeutics: The Ancer to Cancer
Building on 18 years of commercially successful research and development in the field of computational immunology, EpiVax developed Ancer™, a personalized cancer vaccine platform. Ancer is a high-speed, secure, cloud-based commercial platform for processing cancer/normal protein sets, identifying high-quality patient-specific antigens, and designing personalized cancer vaccines. EpiVax promoted this tool as a new venture, EpiVax Therapeutics (EVT), in early 2017 as a wholly owned, investor-backed subsidiary. EVT employs a world-leading technology, developed over 23 years by EpiVax, to design vaccines that activate the body’s T cells to cure or prevent disease in the host. The objective is to become the leading personalized cancer vaccine company, while addressing the major medical needs unmet by other oncology and immuno-oncology treatment approaches. EVT’s unique approach is based on mutated tumor-specific epitopes, termed Neo-epitopes. Genetic profiling is used to generate therapeutic vaccines customized for each cancer patient’s unique tumor load. All of EpiVax’s validated vaccine-design tools, including JanusMatrix, iTEM, and EpiMatrix, are integrated into the Ancer platform leading to quicker more accurate identification of epitopes. Since the subsidiary’s outset, the pipeline has included personalized treatments for bladder cancer, solid tumors, melanoma and most recently, COVID-19.

2017-2020  H7N9 Avian Influenza Immune Engineering and Clinical Trial
H7N9 Avian Influenza Immune Engineering and Clinical Trial
The delayed availability of a vaccine during the 2009 H1N1 influenza pandemic created a sense of urgency to better prepare for the next influenza pandemic, which may have been H7N9 influenza in 2013. When that strain of potentially pandemic influenza emerged, EpiVax received significant funding from the NIH to research a novel vaccine design strategy called “immune engineering” in the context of H7N9 vaccine development. The approach combines EpiVax’s immunoinformatics methods and structure modeling methods of collaborators. The goal of immune engineering is to promote protective antibody responses against H7N9 hemagglutinin (HA) by engineering the whole antigen to include inflammatory T cell epitopes without perturbing the native structure of the protein. Simply put, in two rounds of optimization, EpiVax’s scientists worked to enhance the inflammatory T cell immune response to the antigen by modifying a small number of amino acid residues within the protein sequence. These modifications introduced memory T effector epitopes and removed epitopes that were highly cross-conserved with the human proteome (with regulatory, or immune dampening, potential). Testing in humanized mouse models have demonstrated that this immune engineering technique has increased the immunogenicity of the antigen. Currently, the first-generation vaccine candidate is in headed towards human clinical trials, and promising next-generation vaccine candidates are in development.
2018  Revolutionizing Risk Assessments with PANDA
Revolutionizing Risk Assessments with PANDA
The FDA released guidance which outlined new immunogenicity assessment requirements for synthetic generic peptide drugs, calling for pharmaceutical generic peptide manufacturers to submit information about potential impurities in the drug formulation and to establish that these impurities do not increase the risk of immunogenicity. In response to this guidance, EpiVax developed the Peptide Abbreviated New Drug Application Program, or PANDA. Through a series of steps, PANDA can assess the immunogenicity of the reference listed drug (RLD) and the associated synthetically produced generic peptide products, as well as process-related peptide impurities to evaluate bioequivalence.
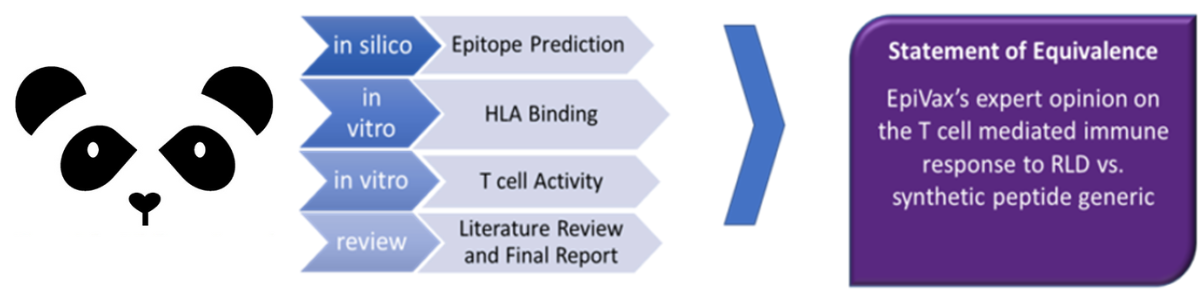
From those steps, our immunogenicity assessment experts compile the results into one comprehensive report, including a detailed description with respect to the predicted immunogenic potential and observed immunogenicity ex vivo of the RLD, the synthetically produced generic peptide product lots, and the process-related peptide impurities. The report also includes opinions and recommendations regarding the immunogenicity of the generic peptide drug and the contribution of the process-related peptide impurities to overall immunogenicity. These formal reports can then be submitted to support the regulatory filing, increasing the likelihood of a favorable review. Over the past few years, EpiVax has seen the PANDA program become increasingly useful for assessing the potential immunogenicity of novel peptide therapeutics. In 2020 alone, EpiVax worked with over fifteen pharmaceutical and biotechnology companies developing both novel and generic peptide therapeutics, applying in silico screening, HLA binding assays, and T cell assays to assess the immunogenic risk of their products utilizing the PANDA approach.
2020  COVID-19 Vaccine
COVID-19 Vaccine
Using well-established and vastly preclinically validated computational vaccinology techniques, EpiVax has helped collaborators develop multiple vaccine designs targeting the SARS-CoV-2 virus and generated an internal epitope-based candidate. It is EpiVax’s hope to bring at least one of these designs forward into human clinical trials to address the COVID-19 global health crisis.

2020  The What-If Machine
The What-If Machine
In the fall of 2020, EpiVax was granted a two-year contract with the FDA to continue partnership with CUBRC, an independent not-for-profit scientific corporation that executes R&D, testing and systems integration programs. During the previous two-year collaboration, EpiVax demonstrated the value of in silico tools and in vitro validation methods for the evaluation of generic peptide drugs and their impurities. The immunogenic risk of two generic peptide drugs were analyzed, applying the concepts outlined in recent FDA draft guidance for ANDAs. Upon the continuation of this collaboration with CUBRC, research carried on, resulting in the development of the What-if Machine (WhIM) – an advanced computational algorithm that performs iterative modifications to each and every amino acid in the sequence of a synthetic peptide drug, entirely in silico. This generates a comprehensive list of all potential impurities that may occur due to deletions, insertions, duplications or side chain modifications at any residue of the active pharmaceutical ingredient (API). The algorithm also ranks those impurities from Highest Risk to Low Risk, flagging those that may be high risk and enabling the FDA to request additional information. It was quickly understood by the EpiVax team that the WhIM has significant potential to contribute to prospective identification of high-risk impurities, allowing for generic peptide drug products to be de-risked early in the development process.

2021  The Tregitope is Licensed
The Tregitope is Licensed
Since the discovery of the Tregitope by Dr. Annie De Groot and Bill Martin in 2008, many more Tregitopes have been validated and Epivax has developed a strong Tregitope patent estate. With a wide range of therapeutic applications, Tregitopes represent a more natural way of inducing tolerance than taking immunosuppressive drugs, and treatment might be initiated early, might be safer, and could have long lasting effects. An exciting step for the continued expansion of the Tregitope program came in 2021, with a commercial licensing agreement between EpiVax and Maruho Co. for the patented technology. Maruho, a Japanese leader in research and development, manufacturing, and commercialization of dermatological products, was able to leverage their Tregitope license to enhance their in-house therapeutic development strategies for autoimmune skin conditions. This agreement reflects the potential for Tregitopes to be used for the treatment of conditions that affect patients globally. Today, the licensing agreement continues as the development of these products is expected to take several years as the final formulation is decided upon. EpiVax is currently licensing Tregitope under 1-year options + license agreements.

2022  Collaborating on a COVID Vaccine
Collaborating on a COVID Vaccine
In collaboration with Intravacc and funded by CEPI (Coalition for Epidemic Preparedness Innovations), EpiVax began the development of universal vaccines for beta-coronaviruses, including SARS-CoV-2 last fall. EpiVax carries out the vaccine epitope selection portion of the collaboration, using the comprehensive in silico vaccine design toolkit, iVAX. Intravacc was awarded $4.8 million to advance their subunit vaccine candidate “Avacc 101”, which is designed for intranasal delivery to provide protective immunity for multiple beta-coronaviruses including SARS-CoV-1, SARS-CoV-2, and MERS-CoV. Avacc 101 uses Intravacc’s Outer Membrane Vesicle technology to deliver vaccine payloads for an effective immune response. As a subgrantee on the project, EpiVax will identify cross-reactive beta-coronavirus T cell epitopes for broad coverage of COVID-19, MERS and SARS Class 1 and Class 2 HLA-restricted epitopes, using iVAX. In addition to vaccine epitope selection, EpiVax will also develop multiple designs, and validate these in silico constructs in vivo with HLA-transgenic mice. The EpiVax team deeply appreciates the opportunity to apply our advanced vaccine design tools to a problem of global significance, living up to our commitment of improving human health everywhere.

2023  CircoMatch
CircoMatch
In result of a recent exclusive partnership with animal health company Zoetis, a unique bioinformatics tool called CircoMatch™ is being developed. CircoMatch™ will allow for better mitigation of the destructive Porcine Circovirus Type 2 (PCV2). PCV2 can cause significant production issues on farms, including suffering and weight loss in growing pigs caused by subclinical infections and reproductive failure in sows. This generates major economic losses to farmers. Current vaccines do help in reducing pig mortality and mitigate economic losses, however, due to continuous viral evolution, these vaccines have varying effectiveness against the many emerging new field strains of PCV2. CircoMatch™ predicts the coverage of varying PCV2 vaccines against field isolates of PCV2. It provides a quick turnaround, and powerful near real-time predictive effectiveness data so swine veterinarians and producers can make informed decisions in their vaccination programs. CircoMatch™ applies EpiVax’s EpiCC tool and PigMatrix algorithm in a custom-built website, which performs a detailed comparison between PCV2 vaccines and circulating field strains of the PCV2 virus. These proprietary computational tools allow for a practical prediction of effectiveness for existing commercial vaccines and may aid in the development of vaccines that are more efficacious across different viral variants.


From 2020 to 2022, EpiVax has been working on “refining and polishing” its many programs, and fine tuning the innovative research progams that it is known for. A new emphasis on Host Cell Proteins has emerged, with funding from the FDA (U01 award) and improved in silico tools for peptides are being developed under contracts to the FDA Office of Generic Drugs.
Lessons learned in the first 25 years? When you are fearless, the ideas are fast and furious, and the innovation never stops. We’ve enjoyed the discoveries AND the collaborations. Stay tuned to find out what new immunoinformatics tools will be invented over the next 25 years of creative science at EpiVax!


