Design superior vaccines through consideration of T cell epitope content:
Consider the importance of T cell epitope-driven immunity.
iVAX™ is an integrated, web-based toolkit for computational vaccine design, available through software licensing agreements, that enables the discovery and optimization of the T cell epitope “triggers” in antigen sequences that are key to activation of protective immune responses. The toolkit enables epitope mapping, antigen selection, immunogen design, and prediction of vaccine efficacy. Facilitating the in silico prediction of immune response to biothreats, emerging infectious diseases, and cancers, these cutting-edge immunoinformatics tools allow for the accelerated design of safer, more effective vaccines for humans and livestock.
The integrated set of tools available in the iVAX™ toolkit stand ready to help vaccine developers deliver better vaccines (of all formats) that consider T cell epitope-driven immunity.
Recent Publication: Better epitope discovery, precision immune engineering, and accelerated vaccine design using immunoinformatics tools. Frontiers in Immunology. 2020.
What can iVAX™ do for you?
Identify Highly Conserved Segments within Viral Strains
Map for T cell Epitopes
Locate Regions of High Epitope Density
Predict Cross-Reactivity of Epitope Sequences and the Human Genome
Create Immunogenic Consensus Sequences
Design a Complete Vaccine Candidate
In the Toolkit:
Map T cell epitopes
Define T cell epitope "hotspots"
Characterize humanness
Securely compare frames with BLAST
Identify conserved segments
Create immunogenic consensus sequences
Identify best arrangement of T cell epitopes
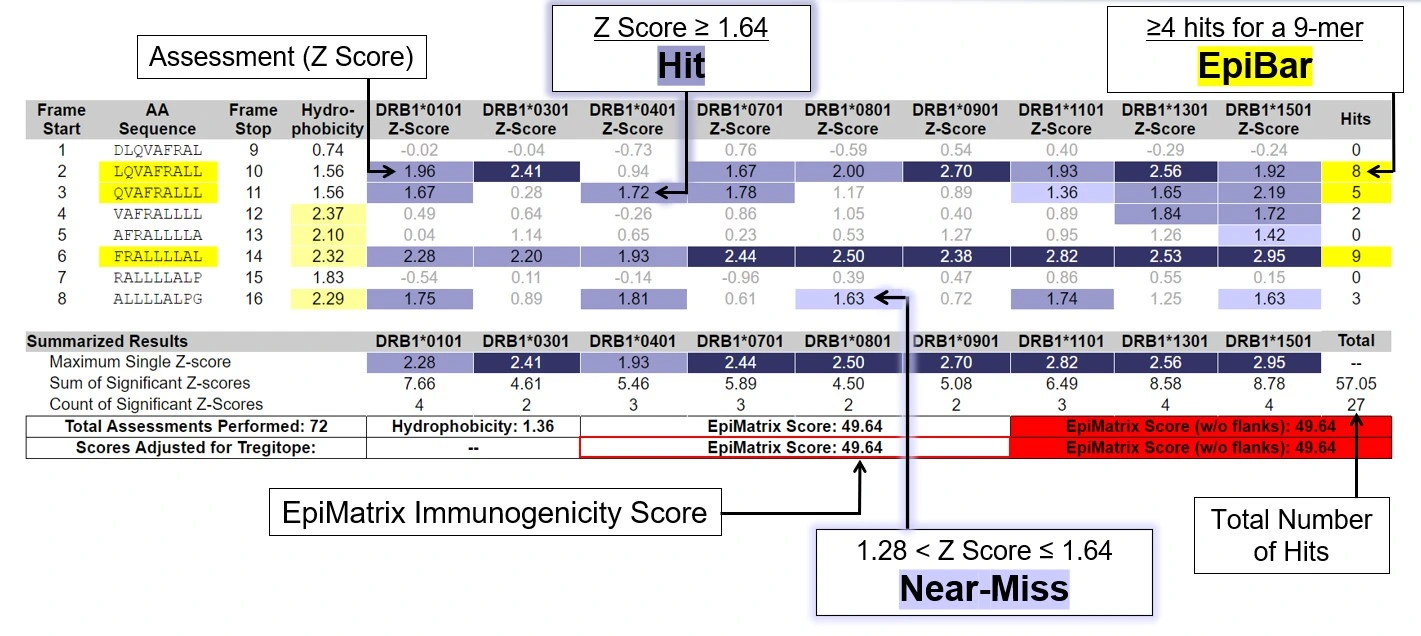
EpiMatrix®
Analyzes overlapping 9-mer frames derived from the target protein sequences and scores them for potential binding affinity against a panel of Class I or Class II HLA alleles: each frame-by-allele assessment that scores highly and is predicted to bind is a putative T cell epitope.
ClustiMer
Screens EpiMatrix output and identifies clusters, or “hotspots”, of 9-mer frames that contain a high density of putative T cell epitopes. These could be regions to focus on for T cell epitope-driven vaccine design and/or antigen optimization.
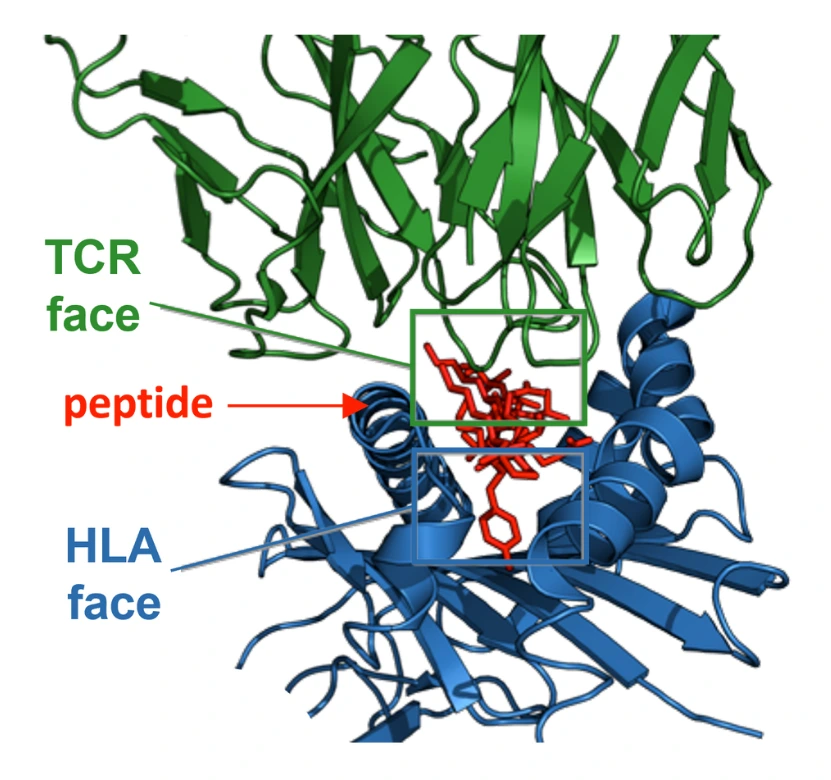
JanusMatrix
Designed to predict the potential for cross reactivity between T cell epitope clusters and the human proteome, based on conservation of TCR-facing residues in their putative HLA ligands. These could be key regions to exclude from a T cell epitope-driven vaccine and/or to reengineer to enhance immunogenicity of an antigen.
BlastiMer
Automates the process of submitting the previously identified frames to BLAST to determine similarities with known proteins.
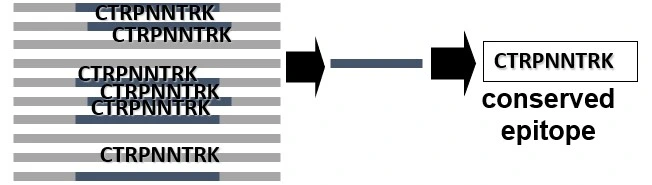
Conservatrix
Parses input sequences into 9-mer strings and identifies those 9-mer sequences that are conserved amongst multiple inputted whole sequences, such as multiple strains of the same pathogen, for even the most mutable of potential vaccine targets.
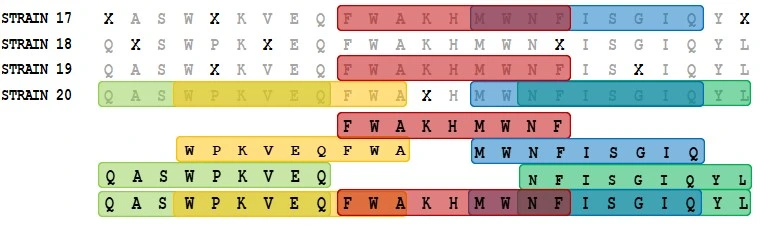
EpiAssembler
Knits together the conserved, immunogenic sequences identified by Conservatrix and EpiMatrix to form highly immunogenic consensus sequences.
VaxCAD
Creates string-of-beads vaccine designs while minimizing deleterious, non-specific junctional T cell epitopes that may be inadvertently created in the process of linking one T cell epitope to another.
Applications:
iVAX™ has been applied to create safer and more effective vaccines in many ways, including by identifying immunogenic T cell epitopes in the development of a T-cell based human multi-epitope Q fever vaccine, designing novel influenza vaccines, identifying cross-conserved T cell epitopes for a malaria vaccine, and analyzing immune responses in clinical vaccine studies. Animal vaccine applications to date have included viral infections of pigs such as swine influenza A, PCV2, and African Swine Fever. “Rapid-Fire” applications for biodefense have included a demonstration project for Lassa Fever and Q fever.
Publications:
T cell epitope engineering: an avian H7N9 influenza vaccine strategy for pandemic preparedness and response. Human Vaccines and Immunotherapeutics. 2018.