Seamlessly integrate immunogenicity screening into your preclinical R&D workflow.
Real-time results for high-throughput screening scenarios.
Proactively manage and mitigate potential immunogenicity challenges.
Build in silico immunogenicity screening into your preclinical R&D process.
EpiVax’s flagship ISPRI™ platform is a comprehensive in silico toolkit for immunogenicity risk assessment and optimization of biologic drugs during drug discovery and preclinical development.
Used for decades by many of the leading global pharmaceutical companies, ISPRI™ is available through software licensing agreements that allow biotech companies to securely and seamlessly integrate immunogenicity screening into their preclinical R&D workflow.
Immunogenicity as a Critical Quality Attribute (CQA) in Drug Discovery
Using ISPRI™, scientists can assess biologic sequences for potential liabilities much earlier by screening a large panel of ‘hits’ for immunogenic potential, allowing for downselection to a smaller panel of leads based on key immunogenicity data points.
Develop a Detailed Immunogenicity Risk Assessment (IRA) Profile
In addition to guiding your hit and lead selection process, ISPRI™ can help you generate a detailed data package summarizing the immunogenic potential of a single candidate.
Improve Promising Candidates Through In Silico
Deimmunization and Humanization
If you have an otherwise promising candidate that is demonstrating unacceptable immunogenicity risk, the OptiMatrix tool in ISPRI™ (shown below) will allow you to quickly model deimmunizing or humanizing changes to mitigate any liabilities.

Conclusion
ISPRI™’s easy-to-use interface delivers real-time results, even in high-throughput screening scenarios involving hundreds to thousands of sequences simultaneously. The ability to understand both the overall and regional immunogenic potential of lead candidates, coupled with the capacity to deimmunize or humanize sequences, positions developers to proactively manage and mitigate potential challenges. With ISPRI™, you’re able to strategically direct R&D efforts, leveraging in silico immunogenicity data to inform your follow-on in vitro and clinical immunogenicity strategy. ISPRI™ can be a catalyst for enhancing the number of viable leads available for development, ultimately de-risking your R&D pipeline and increasing your chances of success.
Modalities
What can ISPRI™ do for you?
- Set the stage with a high-level assessment of sequence risk potential with whole biologic-level scores driven by T cell epitope mapping in a global assessment.
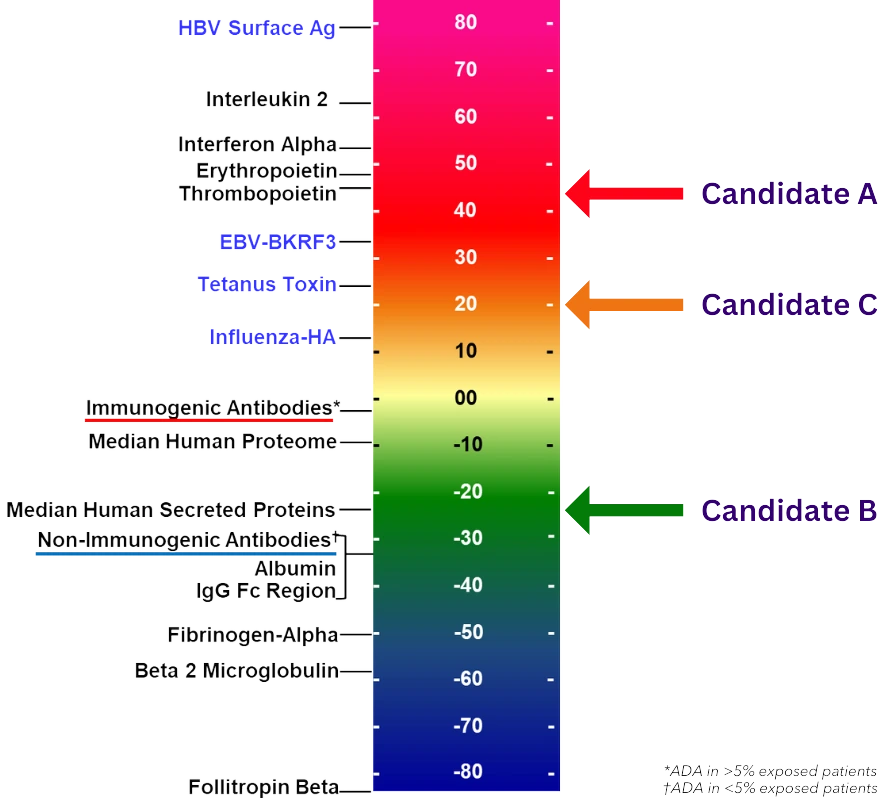
- Rank the overall immunogenic potential of each sequence on a normalized Immunogenicity Scale against similar products with known immunogenicity for easy benchmarking.
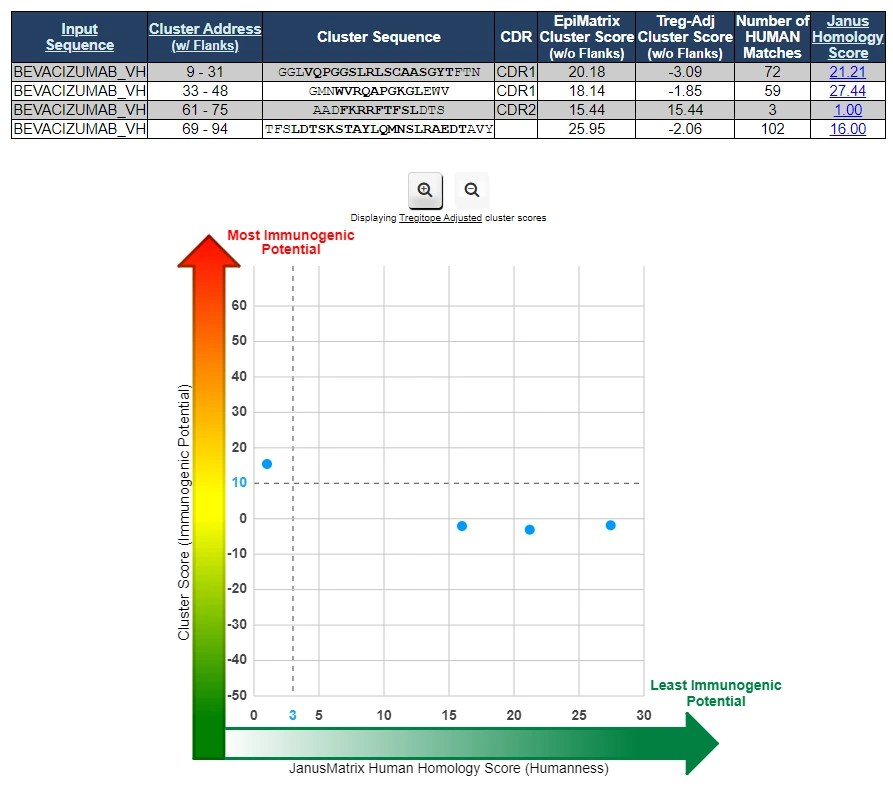
- Dig deeper into sequence analysis with a regional assessment that can uncover sequence hotspots that could be key regions for further investigation and optimization.
- Gain insight into potential tolerance of key regions by comparing them to human epitopes in an advanced homology assessment.
- Securely cross-reference your analyses to published data to better enable your decision making.
- De-risk sequences, if necessary, by strategically introducing point mutations to reduce sequence hotspots.
Recent publication: In silico methods for immunogenicity risk assessment and human homology screening for therapeutic antibodies. mAbs. 2024.
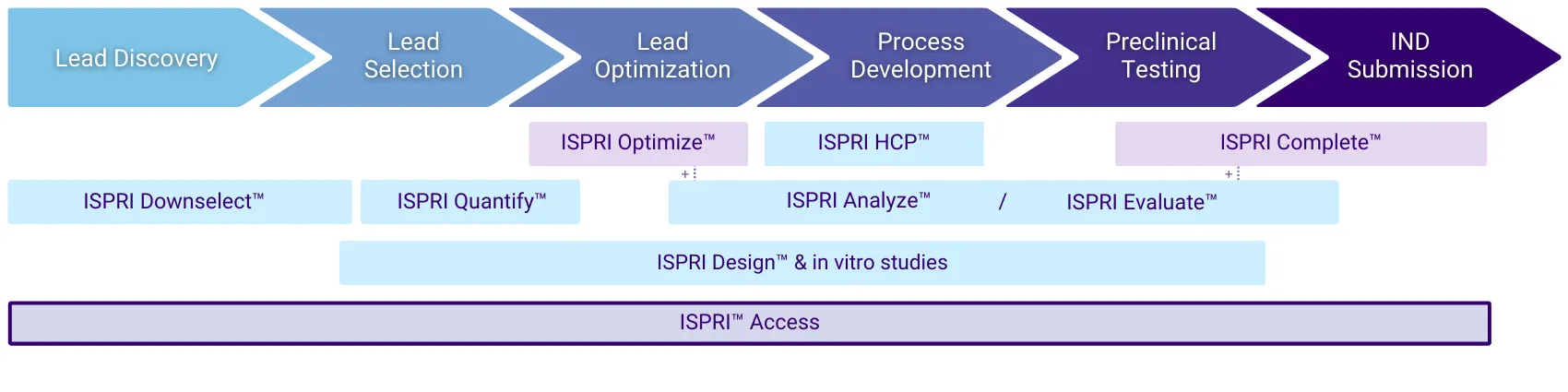
ISPRI™ Use Cases
Lead Discovery
High Throughput Screening of Candidates
- Use immunogenicity as a Key Quality Attribute to help downselect from a large number of hits to a small handfull of leads
- Leverage ISPRI™ Application Programming Interface for quick screening of large numbers of sequences.
Lead Selection
Detailed Assessment of a Single Candidate
- Focus R&D strategies by reducing downstream in vitro and in vivo immunogenicity studies.
- Evaluate in-licensing opportunities.
- Approach regulatory agencies and develop an Integrated Summary of Immunogenicity (ISI) to include in IND filing.
Lead Optimization
Deimmunization of High-Scoring and Non-Human Clusters
- Easily identify and modify immunogenically significant regions, redesigning proteins without introducing new epitopes.
- Iteratively modify individual amino acid residues and see the effect of each modification in real time.
Identify T cell epitopes
Define T cell epitope hotspots
Characterize humanness
Analyze Homology
Deimmunize: premium add-on
EpiMatrix®
What:
Identifies putative effector and regulatory T cell epitopes in your sequence.
How:
Determines HLA binding potential (a prerequisite for T cell activation) and identifies known regulatory T cell epitopes (Tregitopes) against key HLA supertype alleles across a sequence.
Why:
More accurately predict the immunogenic potential of the whole biologic and rank candidates against each other and against benchmark biologics with known immunogenicity.
ClustiMer™
What:
Identifies T cell epitope hotspots in your sequence.
How:
Screens the sequence for regions of T cell epitope density (short peptides predicted to bind to many HLA alleles).
Why:
Dominant T cell epitope clusters can have the potential to drive immune responses in many people, enabling significant immune responses to proteins- in some cases, even those with low global scores. It is key to identify these areas for a better understanding of sequence risk. T cell epitope clusters also make excellent targets for deimmunization.
JanusMatrix™
What:
Analyzes tolerogenic potential of putative T cell epitope content.
How:
Assesses T cell epitope cross-conservation with the human proteome by analyzing TCR-facing residues.
Why:
Characterizing a sequence’s T cell epitope content based on the depth of cross-conservation with human epitopes provides key insight to assess the likelihood that epitopes may be recognized as ‘self’. Epitopes recognized as self may be tolerated by or have tolerogenic effects on the immune system. This characterization is key to a more accurate risk assessment.
Homology Analysis
What:
Performs basic homology analyses of T cell epitope clusters against human and epitope databases.
How:
Cross-references key sequence regions against IEDB and GenBank while keeping your sequences secure.
Why:
Leverage published data to further inform your analysis and potential deimmunization choices.
OptiMatrix™
What:
Deimmunizes high scoring, non-human clusters to reduce immunogenicity or increase tolerance potential.
How:
Selects amino acid substitutions that reduce HLA binding potential or introduce regulatory epitope content in key hotspot regions.
Why:
Reduce sequence risk by removing putative T cell epitope content.
Bring immunogenicity screening in-house:
License ISPRI™!
The EpiVax Roadmap