ISPRI Downselect
Rapidly identify your low-risk candidates.
The ISPRI Downselect™ analysis is a high-level evaluation of the immunogenic potential of a set of biologic therapy candidates. The analysis is typically utilized to rank and inform the selection of high-quality candidates to advance to further development. The analysis, which can be performed on a few to many candidates, is executed by immunoinformatics experts leveraging EpiVax’s proprietary ISPRI in silico toolkit. Results summarizing the T cell epitope content and “humanness” of the whole biologic sequence are provided in a tabulated report, including visual aids that enable the comparison of the immunogenic risk of the sequences of interest (fig. 1). The analysis is accompanied by a discussion with EpiVax experts.
What can ISPRI Downselect™ do for you?
-
Quickly compare the immunogenic potential of small to large sets of biologic sequences.
-
Easily identify low-risk candidates to bring forward in development by leveraging top-tier in silico tools.
-
Save time and money by reducing laboratory work (on average, more than 20-fold)
-
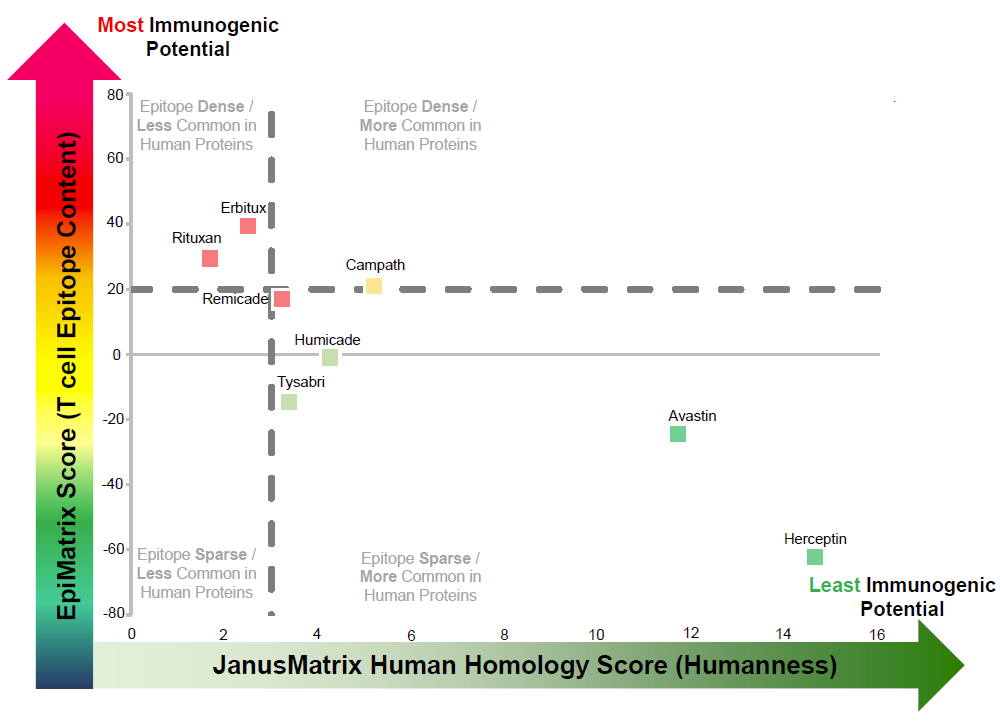
Figure 1. ISPRI Downselect™ data outputs include a quadrant plot like the one to the left, allowing for comparative categorization and visualization of the immunogenic potential of your candidates. T cell epitope content (EpiMatrix) is plotted along the y-axis. A high EpiMatrix score represents an abundance of epitopes and consequently, an elevated immunogenic risk. Human homology (JanusMatrix) is plotted along the x-axis. A high JanusMatrix score represents high cross-conservation with the human proteome and higher likelihood that the therapeutic will be tolerated by the immune system. The plot is divided into quadrants that enable the categorization of sequences into one of four broad immunogenicity risk quadrants: (1) Highest risk – epitope dense, less common in human proteins; (2) moderate-to-high risk – epitope sparse, less common in human proteins; (3) moderate-to-low risk – epitope dense, more common in human proteins; and (4) lowest risk – epitope sparse, more common in human proteins.
Need actionable immunogenicity data?
Learn more about ISPRI Downselect™!
Looking for a comprehensive immunogenicity profile of 1 lead candidate? You need ISPRI Evaluate!
The EpiVax Roadmap