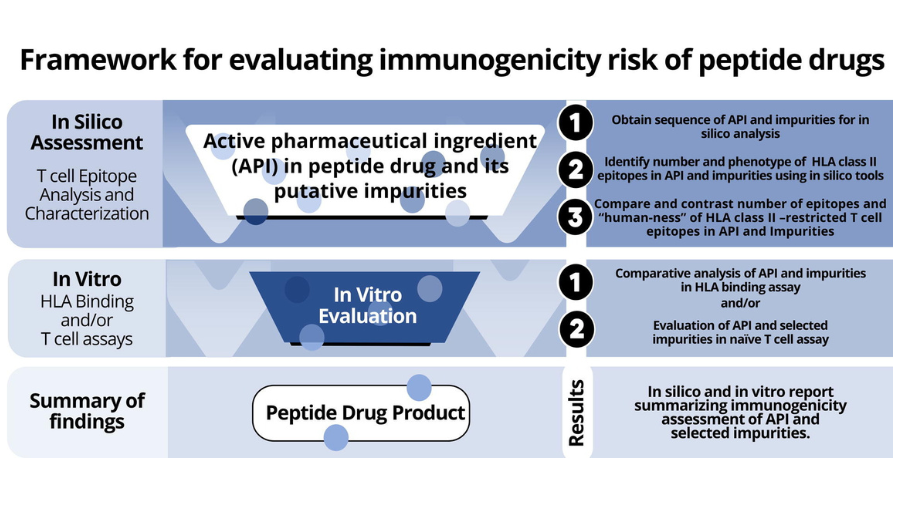
Immunogenicity assessment is key to drug safety. Currently there is no standard approach to the In Silico Immunogenicity Assessment of Therapeutic Peptides, however, the EpiVax team has worked with the FDA to develop several methods, including a validated in silico approach. These methods are collected in an overall in silico and in vitro combination of methods, named PANDA (for peptide Abbreviated New Drug Application) that is carefully aligned with new and updated regulatory guidelines for the accelerated approval of generic peptide drugs.
In the context of generic peptide drugs, the FDA abbreviated new drug application pathway guidance describes how product-specific impurities may arise during the production of generic drugs, even though the drug substance (DS) and the reference drug product (DP) are identical in sequence and bioactivity. The guidance requests that sponsors conduct immunogenicity risk assessment of the impurities using orthogonal (independent) assay methods. The FDA guidance specifically mentions new “T cell epitopes” that may be introduced on a drug product, in peptide impurities, as being a focus of specific concern.
The first orthogonal method used for the ANDA pathway is In Silico Immunogenicity Assessment of Therapeutic Peptides, using immunoinformatics algorithms. A second method involves HLA binding assays. A third method involves in vitro T cell assays. EpiVax has developed state-of-the-art in silico tools and in vitro validation methods for each of these steps in the immunogenicity risk assessment of biologic products. The approach was developed in the course of research carried out under two sequential FDA research contracts with the Office of Generic Drugs (OGD). This approach was recently described in a comprehensive publication authored by EpiVax experts that covers both the in silico and in vitro methods; “Immunogenicity risk assessment of synthetic peptide drugs and their impurities”.
For the In Silico Immunogenicity Assessment of Therapeutic Peptides, EpiVax employs precise, extensively validated algorithms such as EpiMatrix and JanusMatrix (described below). An additional algorithm called the What If Machine (WhIM) has been developed, modelling nearly all impurities that may occur during peptide manufacturing and storage. WhIM generates a list of thousands of theoretical impurities and ranks them from highest to lowest immunogenic potential, using EpiMatrix and JanusMatrix.
EpiMatrix: This algorithm analyzes the immunogenic potential of the amino acid sequence of a given protein or peptide sequence by parsing the sequence into 9-mer peptides. These 9-mers are then assessed for binding potential to a set of HLA “supertype” alleles. For the analysis of generic peptides, the HLA DR alleles used for the analysis include the critical ‘supertype’ alleles, which provide coverage of greater than 95% of the HLA DRB1 types expressed in the global population [1]. Positive EpiMatrix scores indicate abundance of epitopes compared to a random expectation, and consequently an elevated immunogenic risk [2].
JanusMatrix: The JanusMatrix algorithm [3, 4] is applied to identify T cell epitopes that are conserved with epitopes found in the human proteome (“self”). Higher numbers of JanusMatrix-matched human sequences indicate greater potential of being tolerated by the human immune system when compared to predicted epitopes with low JanusMatrix Scores.
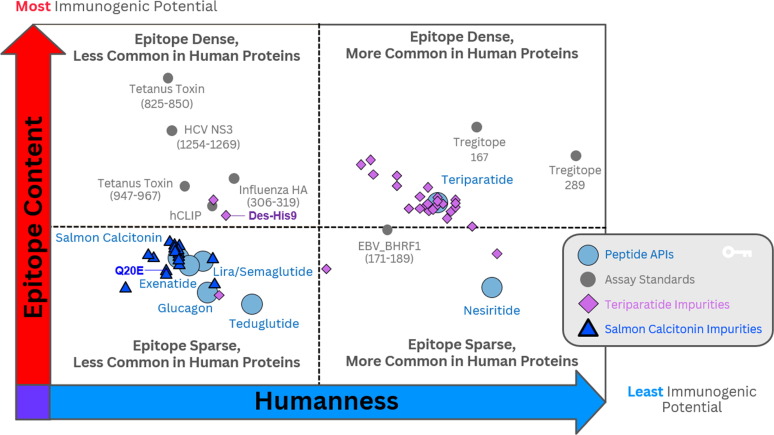
Immunogenicity quadrant analysis categorizes peptides and impurities by immunogenicity risk. EpiMatrix (EMX) and JanusMatrix Human Homology (JMX) scores are plotted for each peptide and impurity. The y-axis represents total epitope content or immunogenic potential. The JanusMatrix score of as given peptide or protein indicates the average depth of coverage (in the human genome) for the HLA binding peptides contained within that sequence.
The predictions performed by EpiMatrix and JanusMatrix have been extensively validated using assays deveoped at EpiVax under FDA contracts HHSF223201810186C and 75F40120C00157. EpiVax’s approach to the in silico and in vitro evaluation of generic peptide drugs has significantly advanced the state of the art in biologics safety and risk assessment and ameliorated the safety of drugs available to the American public.
For more information:
A recent publication on the EpiVax approach to the In Silico Immunogenicity Assessment of Therapeutic Peptides that contain unnatural Amino Acid, published in Frontiers in Immunology by EpiVax immunoinformatics expert Aimee Mattei in 2023
An overview of the industry standard for immunogenicity risk assessment was published by EpiVax founder, former CEO and current CSO, Annie De Groot, with her colleagues across the biotech industry: T-Cell Dependent Immunogenicity of Protein Therapeutics Pre-clinical Assessment and Mitigation–Updated Consensus and Review in 2020