JanusMatrix Analysis
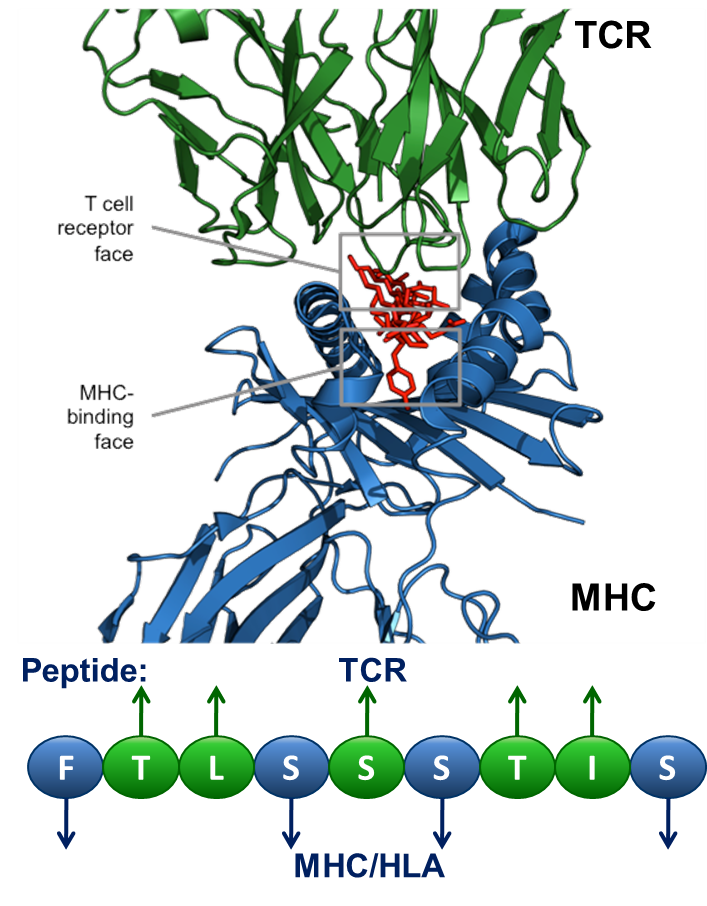
Each HLA ligand has two faces: the HLA-binding face (agretope, blue residues above), and the TCR-interacting face (epitope, green residues above). Predicted ligands with identical epitopes and variant agretopes may stimulate cross reactive T cell responses, providing they bind to the same HLA allele.
The JanusMatrix algorithm, included in the ISPRI toolkit, is a key differentiator of EpiVax’s technology.
JanusMatrix, named after Janus — the Roman god of duality, is used to identify putative epitopes which have the potential to cross-react with the human genome. The JanusMatrix algorithm first considers all the predicted epitopes contained within a given protein sequence and divides each predicted epitope into its constituent agretope and epitope. Each sequence is then screened against a database of human proteins.
Human peptides with a compatible HLA facing agretope (i.e. the agretopes of both the input peptide and its human counterpart are predicted to bind to the same HLA allele) and exactly the same TCR facing epitope are returned. The JanusMatrix Human Homology Score of a given peptide or protein indicates the average depth of coverage within the human genome for the HLA binding peptides contained within that sequence. Therefore, a high JanusMatrix Human Homology Score suggests a bias towards immune tolerance.
In the case of a therapeutic protein, cross conservation between autologous human epitopes and epitopes in the therapeutic may increase the likelihood that such a candidate will be tolerated by the human immune system.
In the case of a vaccine, cross conservation between human epitopes and the antigenic epitopes may indicate that such a candidate utilizes immune camouflage, thereby evading the immune response and making for an ineffective vaccine.
